Back
Introduction: To assess whether radiomic features can predict histopathology after pcRPLND in metastatic non-seminomatous testicular cancer
Methods: Prospective dataset:855 features for 122 patients.The tomography images not used directly but their radiomic features were extracted using the open-source python package pyradiomics. Number of features reduced using Principal Component Analysis to a nal n= 25 principal components. Clinical data:brosis/necrosis (n=57)teratoma (n=48)viable tumor(n=17).Given the strong imbalance among label types, random oversampling applied on the minority class (viable tumor)
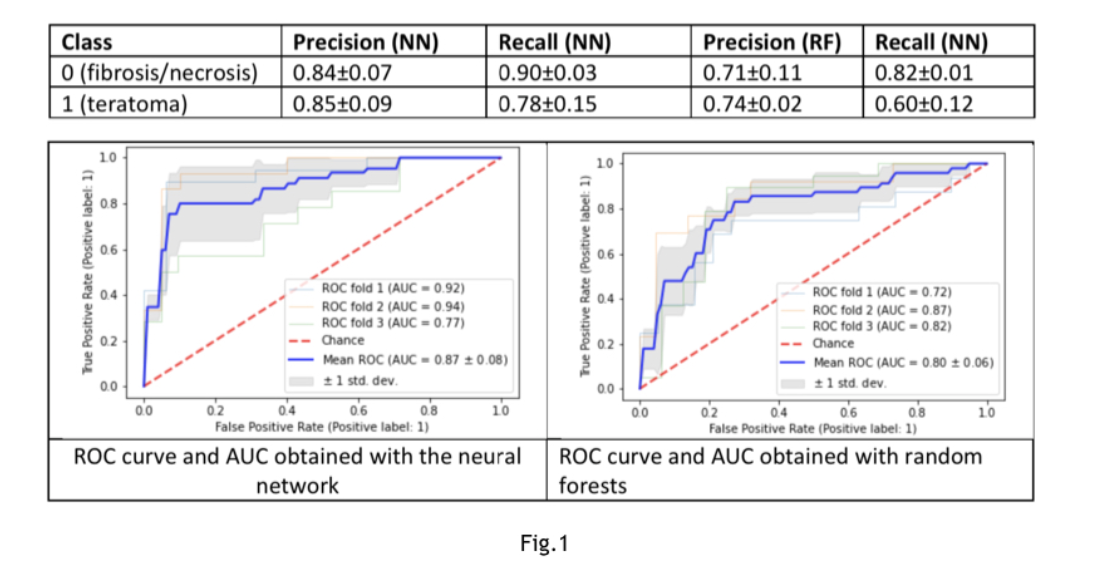
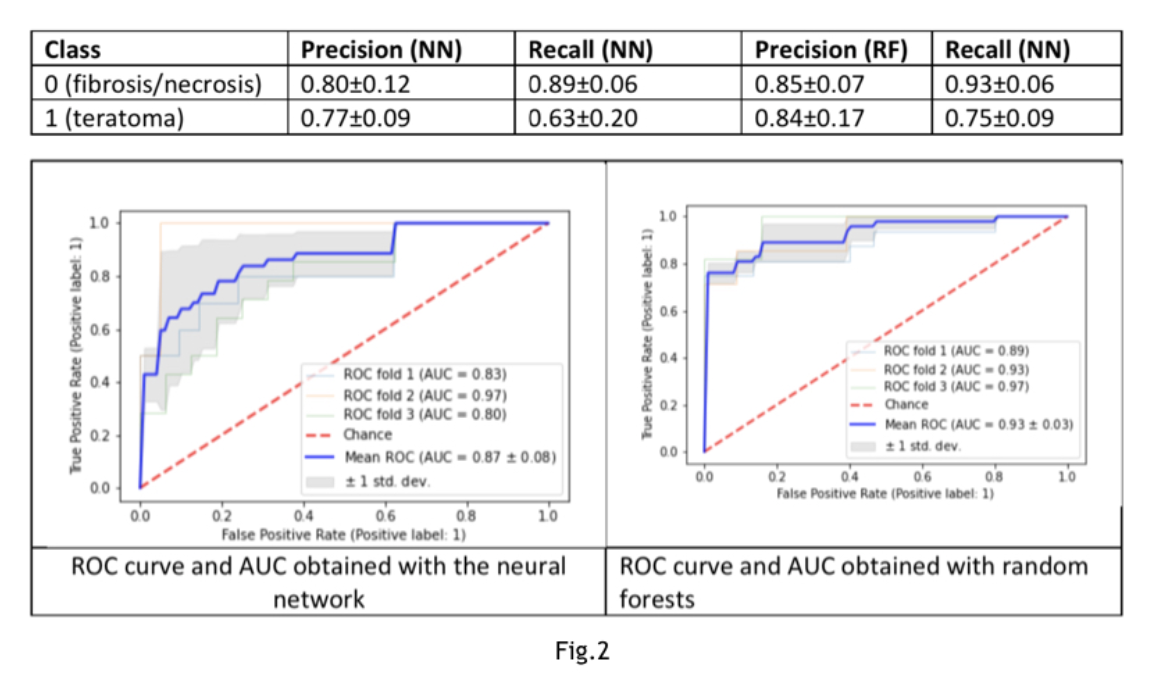
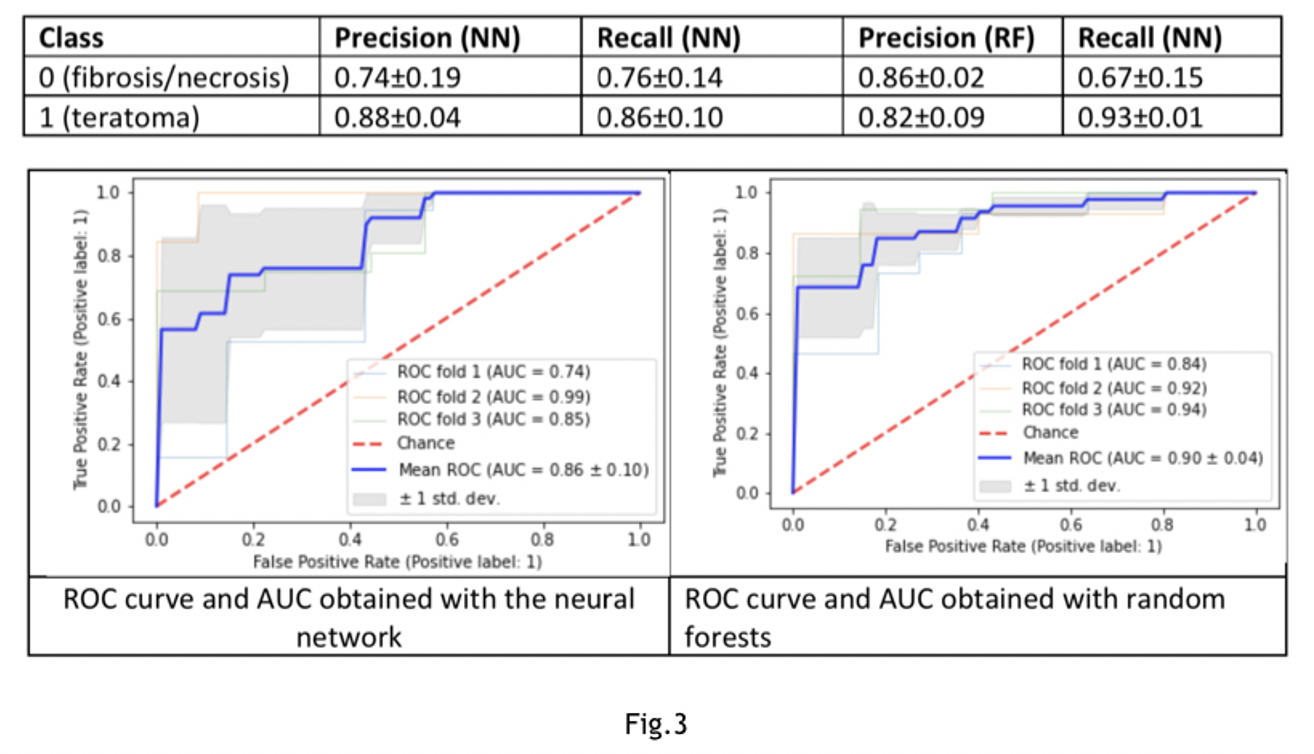
Results: The observations corresponding teratoma(n=48)vs brosis/necrosis (n=57)are 105. Two models compared: neural network (NN) composed of two dense layers of 100 nodes, activation function “relu”, 20% dropout after each layer and a random forests classier (RF). Accuracy obtained with NN:0.85±0.07 with RF:0.72±0.05 (Fig.1). The observations corresponding viable tumors(n=17class1)vs brosis/necrosis (n=57 class0). Accuracy obtained with NN: 0.79±0.01 with RF: 0.79±0.01(Fig.2).The observations corresponding to viable tumors(n=17class0)vs teratoma (n=48class1). A 60% oversampling on the minority class applied. The Accuracy obtained with NN: 0.83±0.09, with RF:0.83±0.07(Fig.3).
Conclusions: Radiomics model is accurate to recognize between teratoma vs tumor viable vs necrosis. Worthwhile tool to reduce overtreatment. SOURCE OF
Funding: None
Moderated Poster Session
Session: MP33: Penile and Testicular Cancer I
MP33-16: Radiomics Model to predict teratoma in metastatic non-seminomatous testicular germ cell tumors
Saturday, April 29, 2023
9:30 AM – 11:30 AM CST
Location: S404C
- AS
Poster Presenter(s)
Introduction: To assess whether radiomic features can predict histopathology after pcRPLND in metastatic non-seminomatous testicular cancer
Methods: Prospective dataset:855 features for 122 patients.The tomography images not used directly but their radiomic features were extracted using the open-source python package pyradiomics. Number of features reduced using Principal Component Analysis to a nal n= 25 principal components. Clinical data:brosis/necrosis (n=57)teratoma (n=48)viable tumor(n=17).Given the strong imbalance among label types, random oversampling applied on the minority class (viable tumor)
Results: The observations corresponding teratoma(n=48)vs brosis/necrosis (n=57)are 105. Two models compared: neural network (NN) composed of two dense layers of 100 nodes, activation function “relu”, 20% dropout after each layer and a random forests classier (RF). Accuracy obtained with NN:0.85±0.07 with RF:0.72±0.05 (Fig.1). The observations corresponding viable tumors(n=17class1)vs brosis/necrosis (n=57 class0). Accuracy obtained with NN: 0.79±0.01 with RF: 0.79±0.01(Fig.2).The observations corresponding to viable tumors(n=17class0)vs teratoma (n=48class1). A 60% oversampling on the minority class applied. The Accuracy obtained with NN: 0.83±0.09, with RF:0.83±0.07(Fig.3).
Conclusions: Radiomics model is accurate to recognize between teratoma vs tumor viable vs necrosis. Worthwhile tool to reduce overtreatment. SOURCE OF
Funding: None



