Back
Ignite Talk
Session: Ignite Session 1A
1406: Integrative Multi-omic Phenotyping in Blood Identifies Molecular Signatures and Candidate Biomarkers of ACPA-negative Rheumatoid Arthritis
Saturday, November 12, 2022
1:45 PM – 1:50 PM Eastern Time
Location: Northern Liberties Stage
- BH
Benjamin Hur, PhD
Mayo Clinic
Rochester, MN, United StatesDisclosure: Disclosure information not submitted.
Ignite Speaker(s)
Benjamin Hur1, Kevin Cunningham2, John Davis1 and Jaeyun Sung1, 1Mayo Clinic, Rochester, MN, 2University of Minnesota, Rochester, MN
Background/Purpose: ACPA detection assays are often used for RA diagnosis due to their high specificity ( >90%). However, current ACPA assays (e.g., anti-CCP2 ELISA) have far lower sensitivity (30–60%), and thus patients can still be clinically diagnosed with RA while testing negative for ACPA (or otherwise known as the "ACPA-negative" RA subgroup). This can make the diagnosis of RA quite challenging for early-stage RA patients, resulting in delays in starting therapeutic interventions. In addition, ACPA-negative RA patients are about 40% of the RA patient population and can show different disease course compared to ACPA-positive patients. To better understand the biomolecular characteristics of ACPA-negative RA given its clinical significance, we performed an integrative multi-omic phenotyping analysis on blood (plasma) samples from RA patients of two different subgroups (i.e., ACPA-negative, ACPA-positive) and healthy controls. Our aims were to identify features specific to ACPA-negative RA and potential biomarkers for ACPA-negative RA diagnostics.
Methods: Global proteomic, metabolomic, and autoantibody profiling were performed on plasma samples from ACPA-negative RA patients (n = 40), sex-/age-matched ACPA-positive RA patients (n = 40), and healthy controls (n = 40) (Fig. 1). A linear regression model was used to identify multi-omics features associated with ACPA-negative RA. ElasticNet was used to infer a multiplex network (from the multi-omics data), which characterizes associations among multi-omic features and clinical conditions in a single analytical framework (Fig. 2A, Steps 1–2). A machine-learning strategy using a network-based feature selection scheme was developed for RA subgroup classification (Fig. 2A, Steps 3–5).
Results: We identified various plasma proteins, metabolites, or autoantibodies associated with RA subgroups. In particular, compared to healthy controls, we identified 23 upregulated and 49 downregulated plasma proteins in ACPA-negative RA; and 25 upregulated and 32 downregulated plasma proteins in ACPA-positive RA. Proteins that were differentially abundant in ACPA-negative RA were found to be enriched in innate immune response and metabolic processes, while proteins that were differentially abundant in ACPA-positive RA were found to be enriched in chemotaxis- and cell migration-related functions. Interestingly, the identified differentially abundant metabolites and autoantibodies were found to support our proteomics results. Finally, our network-based machine-learning classification scheme was able to distinguish ACPA-negative RA patients from healthy controls with 90.0% accuracy, and classify RA patients (of both subgroups) and healthy controls with 86.6% accuracy (Fig. 2B).
Conclusion: We demonstrate the first simultaneous proteomic, metabolomic, and autoantibody phenotyping in the blood of patients with ACPA-negative RA. Our computational analysis identifies multi-omics features that may explain the similarity and differences in biomolecular processes between the ACPA-negative and ACPA-positive subgroups of RA; and suggests potential biomarkers with clinical applicability in blood-based RA diagnostics.
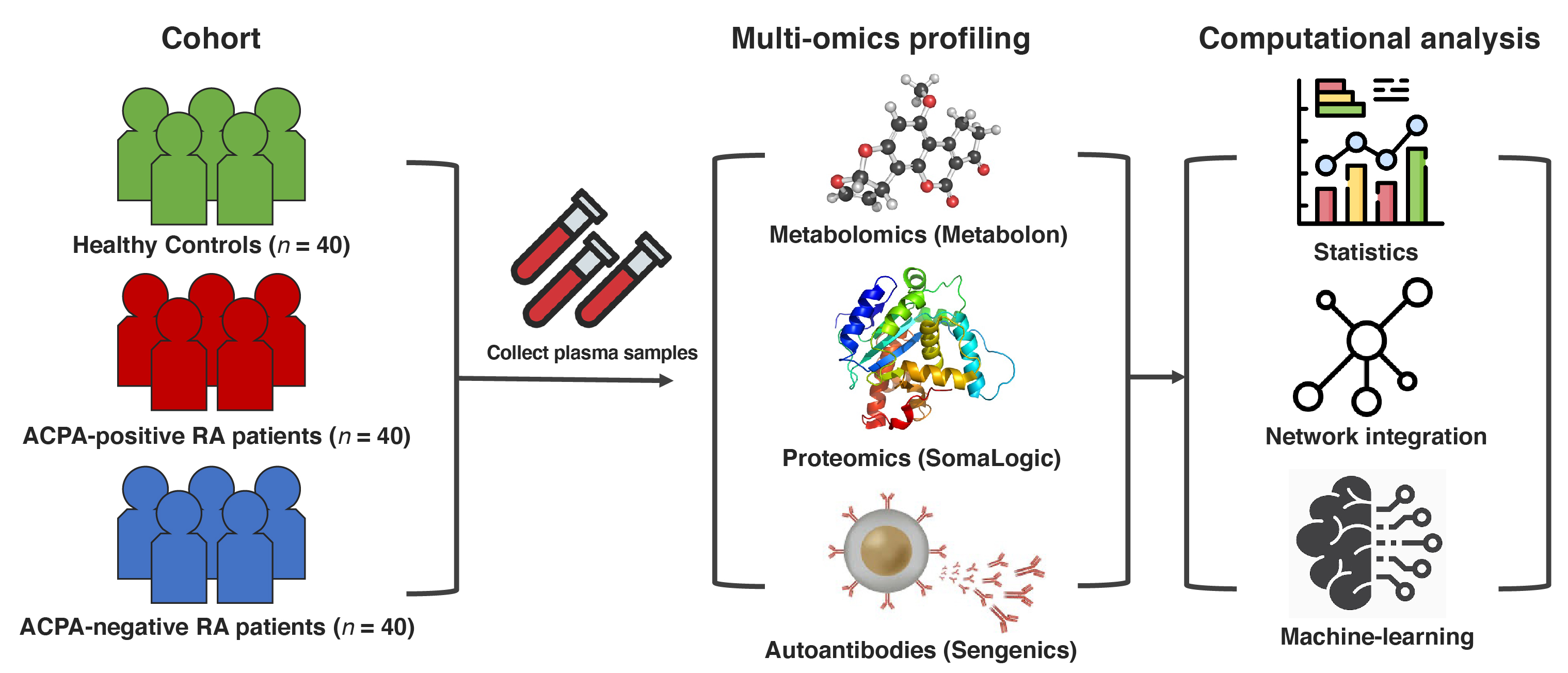 Figure 1. Study design overview and data analysis strategy. Plasma samples were collected from ACPA-negative RA patients (n = 40), sex-/age-matched ACPA-positive RA patients (n = 40), and healthy controls (n = 40). Global proteomic, metabolomic, and autoantibody phenotyping were performed on the plasma samples. On the ensuing multi-omics data, computational analyses were performed to identify biomolecular features specific to ACPA-negative RA and potential computational biomarkers for ACPA-negative RA diagnostics.
Figure 1. Study design overview and data analysis strategy. Plasma samples were collected from ACPA-negative RA patients (n = 40), sex-/age-matched ACPA-positive RA patients (n = 40), and healthy controls (n = 40). Global proteomic, metabolomic, and autoantibody phenotyping were performed on the plasma samples. On the ensuing multi-omics data, computational analyses were performed to identify biomolecular features specific to ACPA-negative RA and potential computational biomarkers for ACPA-negative RA diagnostics.
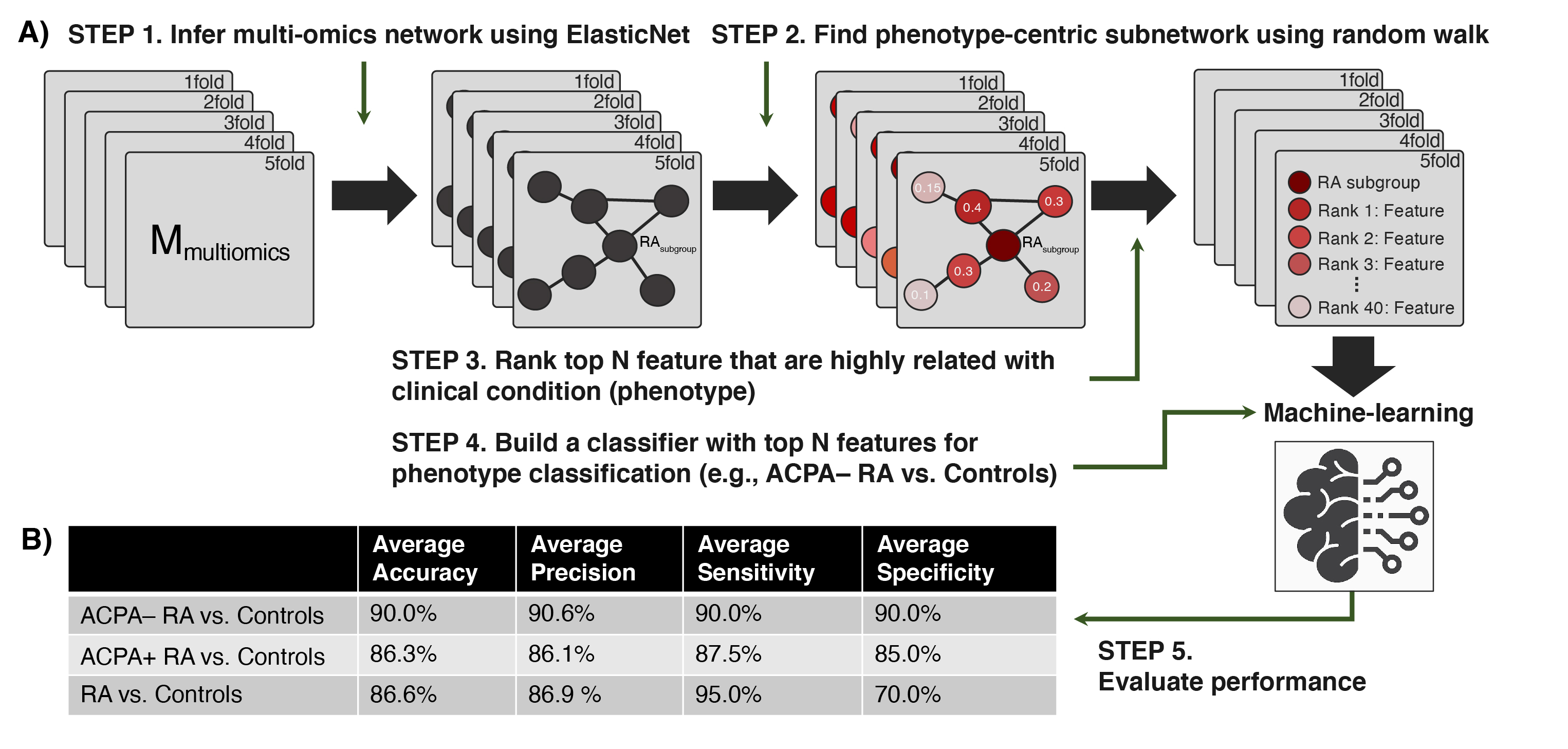 Figure 2. Performance of our network-driven, integrative multi-omics biomarker discovery approach was evaluated in 5-fold cross-validation. (A) A machine-learning framework that (i) infers a multiplex network from multi-omics data, (ii) identifies a subset of the multiplex network (a subnetwork) that includes only the features most associated with an RA subgroup, and (iii) uses the top N features to design classifiers for RA. (B) Average accuracy, precision, sensitivity, and specificity in each classification scenario. Of note, our machine-learning strategy obtained 90.0% (or slightly higher) results in all four metrics when distinguishing ACPA-negative RA from controls.
Figure 2. Performance of our network-driven, integrative multi-omics biomarker discovery approach was evaluated in 5-fold cross-validation. (A) A machine-learning framework that (i) infers a multiplex network from multi-omics data, (ii) identifies a subset of the multiplex network (a subnetwork) that includes only the features most associated with an RA subgroup, and (iii) uses the top N features to design classifiers for RA. (B) Average accuracy, precision, sensitivity, and specificity in each classification scenario. Of note, our machine-learning strategy obtained 90.0% (or slightly higher) results in all four metrics when distinguishing ACPA-negative RA from controls.
Disclosures: B. Hur, None; K. Cunningham, None; J. Davis, Pfizer; J. Sung, None.
Background/Purpose: ACPA detection assays are often used for RA diagnosis due to their high specificity ( >90%). However, current ACPA assays (e.g., anti-CCP2 ELISA) have far lower sensitivity (30–60%), and thus patients can still be clinically diagnosed with RA while testing negative for ACPA (or otherwise known as the "ACPA-negative" RA subgroup). This can make the diagnosis of RA quite challenging for early-stage RA patients, resulting in delays in starting therapeutic interventions. In addition, ACPA-negative RA patients are about 40% of the RA patient population and can show different disease course compared to ACPA-positive patients. To better understand the biomolecular characteristics of ACPA-negative RA given its clinical significance, we performed an integrative multi-omic phenotyping analysis on blood (plasma) samples from RA patients of two different subgroups (i.e., ACPA-negative, ACPA-positive) and healthy controls. Our aims were to identify features specific to ACPA-negative RA and potential biomarkers for ACPA-negative RA diagnostics.
Methods: Global proteomic, metabolomic, and autoantibody profiling were performed on plasma samples from ACPA-negative RA patients (n = 40), sex-/age-matched ACPA-positive RA patients (n = 40), and healthy controls (n = 40) (Fig. 1). A linear regression model was used to identify multi-omics features associated with ACPA-negative RA. ElasticNet was used to infer a multiplex network (from the multi-omics data), which characterizes associations among multi-omic features and clinical conditions in a single analytical framework (Fig. 2A, Steps 1–2). A machine-learning strategy using a network-based feature selection scheme was developed for RA subgroup classification (Fig. 2A, Steps 3–5).
Results: We identified various plasma proteins, metabolites, or autoantibodies associated with RA subgroups. In particular, compared to healthy controls, we identified 23 upregulated and 49 downregulated plasma proteins in ACPA-negative RA; and 25 upregulated and 32 downregulated plasma proteins in ACPA-positive RA. Proteins that were differentially abundant in ACPA-negative RA were found to be enriched in innate immune response and metabolic processes, while proteins that were differentially abundant in ACPA-positive RA were found to be enriched in chemotaxis- and cell migration-related functions. Interestingly, the identified differentially abundant metabolites and autoantibodies were found to support our proteomics results. Finally, our network-based machine-learning classification scheme was able to distinguish ACPA-negative RA patients from healthy controls with 90.0% accuracy, and classify RA patients (of both subgroups) and healthy controls with 86.6% accuracy (Fig. 2B).
Conclusion: We demonstrate the first simultaneous proteomic, metabolomic, and autoantibody phenotyping in the blood of patients with ACPA-negative RA. Our computational analysis identifies multi-omics features that may explain the similarity and differences in biomolecular processes between the ACPA-negative and ACPA-positive subgroups of RA; and suggests potential biomarkers with clinical applicability in blood-based RA diagnostics.
 Figure 1. Study design overview and data analysis strategy. Plasma samples were collected from ACPA-negative RA patients (n = 40), sex-/age-matched ACPA-positive RA patients (n = 40), and healthy controls (n = 40). Global proteomic, metabolomic, and autoantibody phenotyping were performed on the plasma samples. On the ensuing multi-omics data, computational analyses were performed to identify biomolecular features specific to ACPA-negative RA and potential computational biomarkers for ACPA-negative RA diagnostics.
Figure 1. Study design overview and data analysis strategy. Plasma samples were collected from ACPA-negative RA patients (n = 40), sex-/age-matched ACPA-positive RA patients (n = 40), and healthy controls (n = 40). Global proteomic, metabolomic, and autoantibody phenotyping were performed on the plasma samples. On the ensuing multi-omics data, computational analyses were performed to identify biomolecular features specific to ACPA-negative RA and potential computational biomarkers for ACPA-negative RA diagnostics. Figure 2. Performance of our network-driven, integrative multi-omics biomarker discovery approach was evaluated in 5-fold cross-validation. (A) A machine-learning framework that (i) infers a multiplex network from multi-omics data, (ii) identifies a subset of the multiplex network (a subnetwork) that includes only the features most associated with an RA subgroup, and (iii) uses the top N features to design classifiers for RA. (B) Average accuracy, precision, sensitivity, and specificity in each classification scenario. Of note, our machine-learning strategy obtained 90.0% (or slightly higher) results in all four metrics when distinguishing ACPA-negative RA from controls.
Figure 2. Performance of our network-driven, integrative multi-omics biomarker discovery approach was evaluated in 5-fold cross-validation. (A) A machine-learning framework that (i) infers a multiplex network from multi-omics data, (ii) identifies a subset of the multiplex network (a subnetwork) that includes only the features most associated with an RA subgroup, and (iii) uses the top N features to design classifiers for RA. (B) Average accuracy, precision, sensitivity, and specificity in each classification scenario. Of note, our machine-learning strategy obtained 90.0% (or slightly higher) results in all four metrics when distinguishing ACPA-negative RA from controls.Disclosures: B. Hur, None; K. Cunningham, None; J. Davis, Pfizer; J. Sung, None.

